Open Models
We believe that shared, transparent, and fully reproducible models will strengthen the confidence and qualification of these models, thereby extending model-informed decision support throughout therapeutic development.
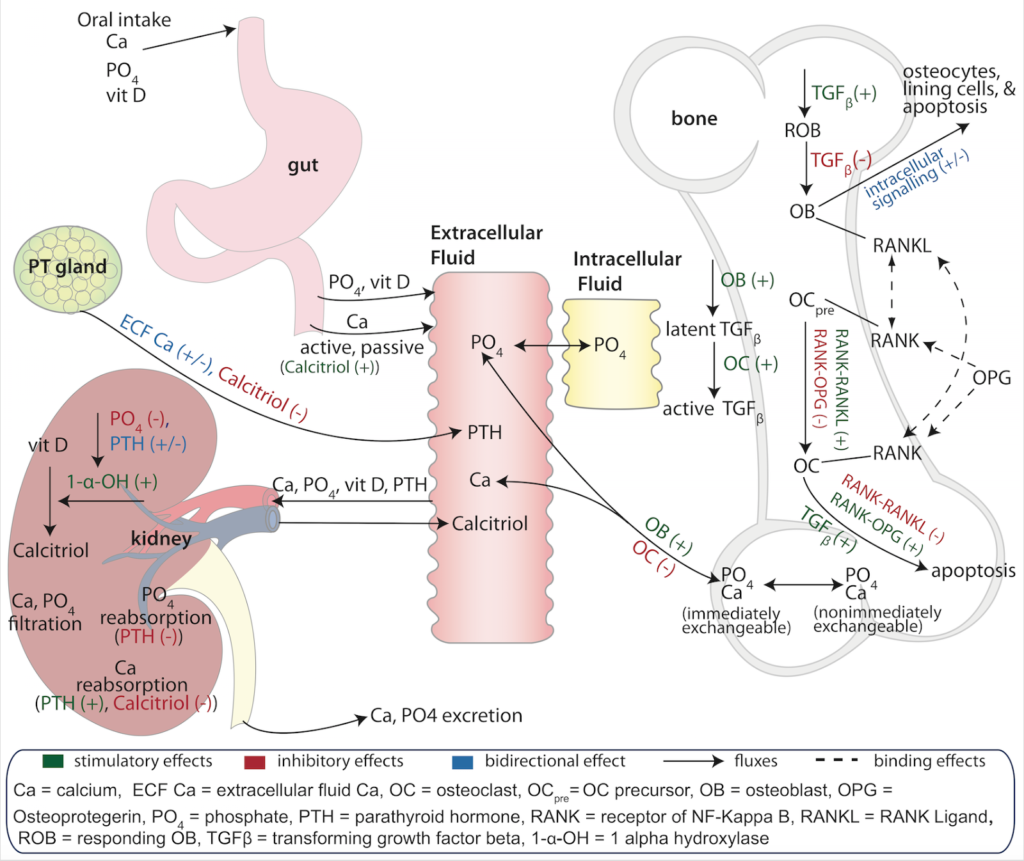
The Bone Health and Mineral Model
We are pleased to announce the free, public release of the Bone Health and Mineral Model! Visit Github to download directly today!
The model was originally developed to describe the bone marker changes associated with denosumab administration from a then ongoing clinical trial. Associated changes in serum calcium and PTH were also considered of interest at the time and so justified the development of a ‘systems’ model that included bone remodeling and bone mineral (calcium and phosphate) homeostatic mechanisms. Other therapeutics (e.g., teriparatide) and disease states (kidney failure, parathyroid-related abnormalities) were also considered at the time to further inform the parameterization and estimation of the model (Peterson and Riggs, Bone 2010).
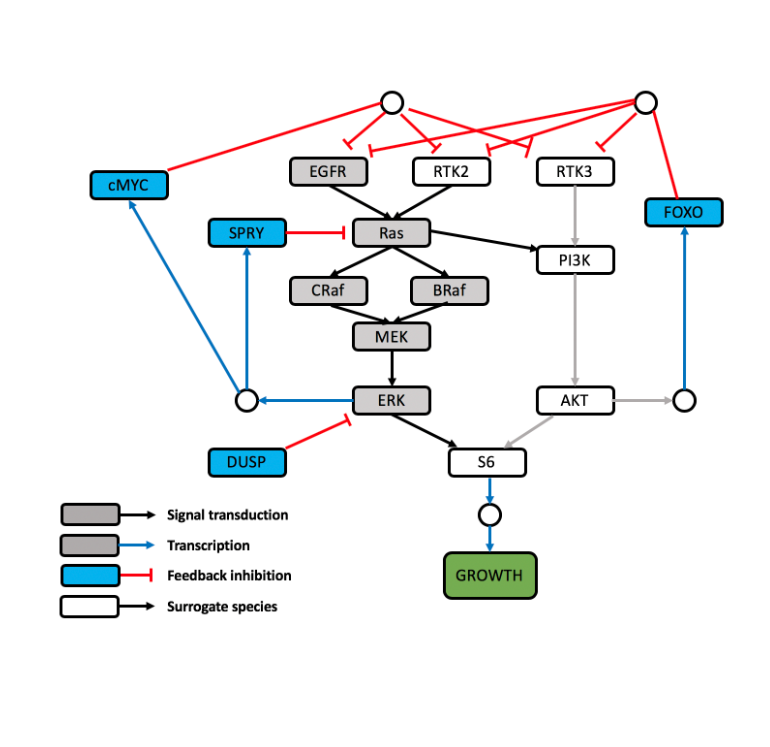
Mitogen‐Activated Protein Kinase (MAPK) QSP Model
A QSP model of the mitogen‐activated protein kinase (MAPK) model (Kirouac et al., NPJ Syst Biol Appl. 2017) was translated for open-source application in R using the mrgsolve package (Elmokadem et al., CPT:PSP 2019). Kirouac et al. were interested in the clinical responses to therapy in colorectal cancer caused by a V600E/K mutation in the cytosolic kinase BRAF, which renders the kinase constitutively active and hence activates the downstream MEK and extracellular‐signal‐regulated kinase (ERK) signaling. BRAF and MEK inhibitors were found to be effective in V600E mutant melanoma, though these agents showed only a moderate effect in colorectal cancer. The purpose of the QSP model was to build a signaling cascade for MAPK to assess the effect of introducing an ERK inhibitor as a monotherapy or in combination regimens with other agents on tumor size in colorectal cancer.
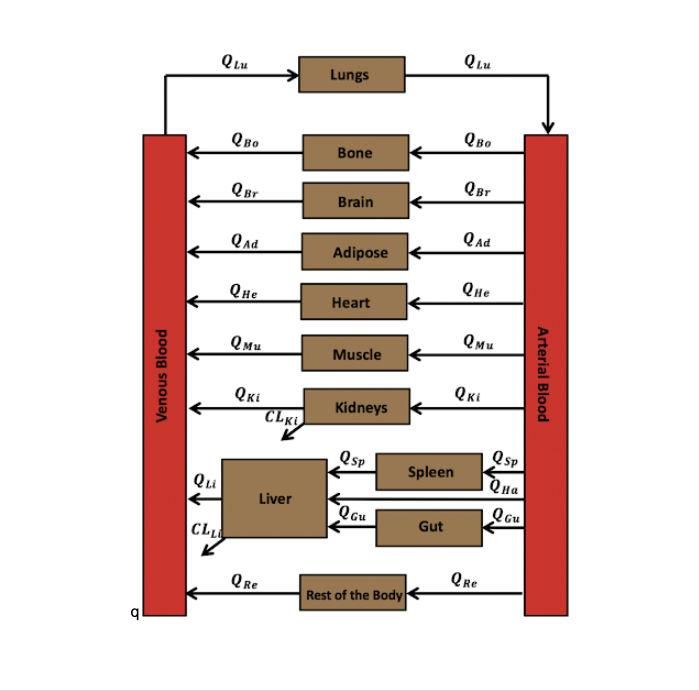
Voriconazole PBPK model implemented in R with mrgsolve
A previously developed voriconazole PBPK model (Zane and Thakker, Clin Pharmacokinet. 2014) was implemented in R using the mrgsolve package as an open-source demonstration of a PBPK platform (Elmokadem et al., CPT:PSP 2019). The PBPK model was developed by Zane and Thakker with the intent of providing a mechanistic explanation for the difference in voriconazole PK between adults and children. These Zane and Thakker results have been recapitulated in the Elmokadem et al., CPT:PSP 2019 and an additional example of parameter optimization was subsequently added to the open-source code repository.